Extracting Parasite Interaction Data from a Scientific Paper using Google Sheets
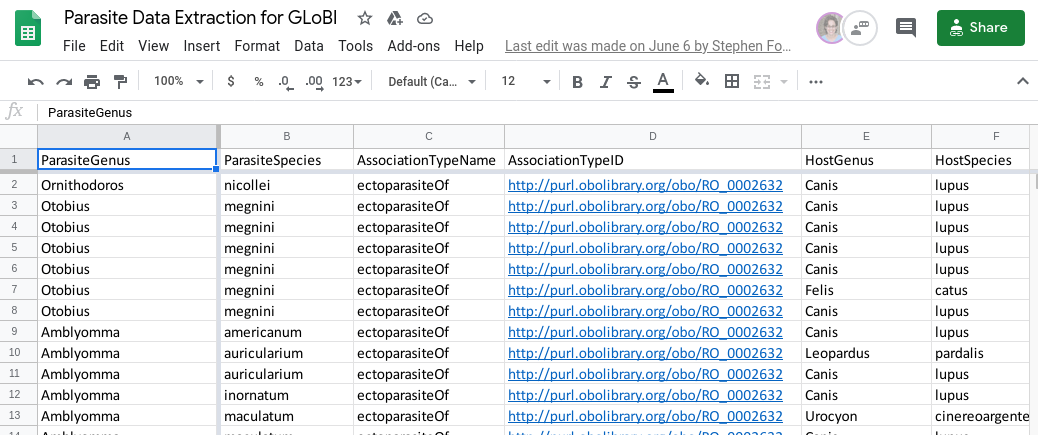
Figure 1. An example of the Google Sheets spreadsheet used for transcribing data. GloBI accessed the transcribed data set through open source scripts accessible on GitHub. Once every other day the data was indexed into GloBI data products.
During my senior year at Texas A&M University, I had the privilege of participating in a data transcription project. The goal of the project was to make parasite data generally more accessible to everyone. My task was to gather data from a host-parasite list from a scientific paper (Light et al. 2019) and prepare it for upload into GloBI by entering information into specific fields on a Google Sheet. Google Sheets was chosen because multiple people were working on this project and it was an efficient way to work simultaneously. I was transcribing the data of records of different ectoparasites (parasites found on the outside of the host body, such as fleas, ticks, mites, and lice) found on rodents across Mexico. For example, Amaradix euphorbia (flea), Otobius megnini (the spinose ear tick), Ameroseius bassolsae (mite), and Hoplopleura hirsuta (a sucking louse). Transcribing data was not a terribly difficult task. The challenge was how quickly I could transcribe the data points from the paper to the Google Sheet.
In my case, each data point consisted of the mammalian ectoparasite, its taxonomy, the host, the taxonomy of the host, the locality, and the natural history collection where the specimen is located (if that information was available). The Google Sheet spreadsheet also included cells that identified the proper taxonomic authorities such as the parasite authority, the host authority, and the association reference.

Figure 2. A spinose ear tick (Otobius megnini), one of the many mammalian parasites that were the subjects of this project. (Lindström, A. 2017).
My goal while transcribing this data was to be as efficient as possible. I did this by copying taxonomy and other information that was the same for multiple rows down about 10 or 20 cells and then hiding those columns so there was less cells I needed to fill in for each row. This process allowed the work to go by much quicker as I only needed to input the genus, species, locality and the home collection of each host-parasite association.
Another challenge I experienced was due to the monotony of the work. It was easy for my mind to slip and make simple mistakes. For example, when I attempted to hide a few columns I accidently erased them and did not realize it until I had already filled in another couple dozen data points. Anyone that has done a lot of work with spreadsheets has probably encountered a problem like this and it can be frustrating and worrisome. Fortunately, the Google Sheet restoration tools allowed me to repair my mistake and maintain the work I had done by copying it to another spreadsheet while restoring the original. To avoid making mistakes like that I worked in short hour-long bursts with breaks in between. This helped significantly by increasing my efficiency and accuracy.
The biggest lesson I learned from working on this project was the importance of giving your brain a break, especially with a monotonous task such as transcribing. I find that if I worked for too long, the mind will look to make shortcuts, which usually leads to mistakes. Taking some time to break apart the work helped keep my mind focused which in turn kept my work accurate. The experience was quite enjoyable. Even though the task was simple, I was satisfied by the work I had done. It was a wonderful privilege to assist in the congregation of demographic data for GloBI, and I am grateful for Dr. Light and her lab allowing me to work on this project.
References
Light, J.E. et al. 2020. Checklist of ectoparasites of cricetid and heteromyid rodents in México. Therya Volume 11 (1): 79-136. doi:10.12933/therya-20-785
Lindström, A., Lindström, J. . 2017. First report of spinose ear tick, Otobius megnini (Acari, Argasidae), in Sweden. Exp Appl Acarol 72, 179–181 . doi:10.1007/s10493-017-0139-5


