The Big Bee network is a project to create and openly share trait information about global bees. This status page shows the current state of integration between Big Bee affiliated collections and GloBI.
ask a question about the project / contribute data
events / collections status / integration profiles / DwC-A guidelines / translation tables / supported biotic interaction terms
Events
15 Sept 2021 - Formal Start of NSF funded projects related to Collaborative Research: Digitization TCN: Extending Anthophila research through image and trait digitization (Big-Bee) including NSF DBI 2102006, NSF DBI 2101929, NSF DBI 2101908, NSF DBI 2101876, NSF DBI 2101875, NSF DBI 2101851, NSF DBI 2101345, NSF DBI 2101913, NSF DBI 2101891, and , NSF DBI 2101850
8 Oct 2021 - Publication of Image Corpus: Cheadle Center for Biodiversity and Ecological Restoration, University of California Santa Barbara. (2021). UC Santa Barbara Invertebrate Zoology Collection (UCSB-IZC) Data Archive and Biodiversity Dataset Graph (0.1) [Data set]. Zenodo. https://doi.org/10.5281/zenodo.5557670 https://archive.org/details/preston-ucsb-izc hash://sha256/d5eb492d3e0304afadcc85f968de1e23042479ad670a5819cee00f2c2c277f36
18 Oct 2021 - Big Bee poster presented by Katja Seltmann at TDWG 2021: Seltmann, K., Allen, J., Brown, B. V, Carper, A., Engel, M. S, Franz, N., et al. (2021). Announcing Big-Bee: An initiative to promote understanding of bees through image and trait digitization. Biodiversity Information Science and Standards, 5(e74037). doi:10.3897/biss.5.74037 Retrieved from https://escholarship.org/uc/item/0937b5gp
28 Oct 2021 - Tara Duggan. (2021). To help fight loss of bees, California Academy of Sciences to share 180,000-strong collection online. San Francisco Chronicle. Retrieved from https://www.sfchronicle.com/climate/article/To-help-fight-loss-of-bees-California-Academy-of-16570034.php on 2 Nov 2021. tweet.
4 Nov 2021 - Publication of Image Corpus: Cheadle Center for Biodiversity and Ecological Restoration, University of California Santa Barbara. (2021). UC Santa Barbara Invertebrate Zoology Collection (UCSB-IZC) Data Archive and Biodiversity Dataset Graph (0.2) [Data set]. Zenodo. https://doi.org/10.5281/zenodo.5660088 hash://sha256/80c0f5fc598be1446d23c95141e87880c9e53773cb2e0b5b54cb57a8ea00b20c https://archive.org/details/preston-ucsb-izc/
29 Nov 2021 - Publication of Big Bee Interaction Data: Seltmann, Katja, & Poelen, Jorrit. (2021). Tab and comma delimited versions of Discover Life bee species guide and world checklist (Hymenoptera: Apoidea: Anthophila) (v55.1) [Data set]. Zenodo. https://doi.org/10.5281/zenodo.5738043.
28 Jan 2022 - Publication of Big Bee Interaction Data: Seltmann, Katja, & Poelen, Jorrit. (2021). Tab and comma delimited versions of Discover Life bee species guide and world checklist (Hymenoptera: Apoidea: Anthophila) (v55.1) [Data set]. Zenodo. https://doi.org/10.5281/zenodo.5915003.
29 Apr 2022 - Publication of Big Bee Interaction Data: Seltmann, Katja C., & Poelen, Jorrit H. (2023). Big Bee indexed biotic interactions and review summary (0.3) [Data set]. Zenodo. https://doi.org/10.5281/zenodo.5915012.
19 May 2022 - Publication of global bee interaction data from GloBI: Katja C. Seltmann. (2022). Global Bee Interaction Data (v1.0.1) [Data set]. Zenodo. https://doi.org/10.5281/zenodo.6564738.
27 Jul 2022 - Publication of Big Bee Interaction Data: Seltmann, Katja C., Poelen, Jorrit H., Allen, Julie, Eldredge, K. Taro, Engel, Michael, Gonzalez, Victor, & Knowles, L. Lacey. (2022). Big Bee indexed biotic interactions and review summary (0.4.0) [Data set]. Zenodo. https://doi.org/10.5281/zenodo.6950082.
26 September 2022 - Publications of Big Bee Images: Big Bee Community, Poelen, Jorrit H., & Seltmann, Katja. (2022). Xylocopa sonorina - UCSB-IZC00012194 - Bee Library - 73e389aa-5886-4c48-8778-ba8932d1bd7e hash://sha256/96bfde1efa599e0e8e61de18b14d61dd308737f684950e4079c04e9bc0f33958 hash://md5/4940f68c84cffa4412f7ffb98bb255bd (0.0.3) [Data set]. Zenodo. https://doi.org/10.5281/zenodo.7114665 .
28 September 2022 - Publication of Seltmann, K. (2022). Extended Thoughts about the Extended Specimen. UC Santa Barbara: Cheadle Center for Biodiversity and Ecological Restoration. Retrieved from https://escholarship.org/uc/item/0g99h7kf. mp4
7 November 2022 - Publication of: Katja C. Seltmann, & Global Biotic Interaction Community. (2022). Global Bee Interaction Data (v2.02) [Data set]. Zenodo. 10.5281/zenodo.7315159
4 January 2023 - Publication of: Baskauf SJ, Girón Duque JC, Nielsen M, Cobb NS, Singer R, Seltmann KC, Kachian Z, Pérez M, Agosti D, Klompen AML (2023) Implementation Experience Report for Controlled Vocabularies Used with the Audubon Core Terms subjectPart and subjectOrientation. Biodiversity Information Science and Standards 7: e94188. https://doi.org/10.3897/biss.7.94188 .
6 January 2023 - Publication of: Seltmann, Katja, Poelen, Jorrit H., UCSB Natural History Collections, & Big Bee Community. (2023). Halictus ligatus Halictidae UCSB-IZC00036803 https://library.big-bee.net/portal/collections/individual/index.php?occid=72096 Male - Bee Library - 73e389aa-5886-4c48-8778-ba8932d1bd7e hash://sha256/3a178bd2f588c35eb9729309d1baa769ea819bcd5bb5142c2de208dd8e299d1a hash://md5/88a7c9b9b2df777a17e707aab635b3ef [Data set]. Zenodo. https://doi.org/10.5281/zenodo.7510912 . 
18 January 2023 - Miller, JT, Poelen, Jorrit, & Seltmann, Katja. (2023). Big Bee Name Alignment Workshop. Zenodo. 10.5281/zenodo.7829969. See also workshop webpage and versioned workshop resources.
31 January 2023 Publication of: Seltmann, Katja C., Poelen, Jorrit H., Allen, Julie, Eldredge, K. Taro, Engel, Michael, Gonzalez, Victor, Knowles, L. Lacey, & Horsley, Pam. (2023). Big Bee indexed biotic interactions and review summary (0.7.0) [Data set]. Zenodo. doi:10.5281/zenodo.7865367
28 February 2023 Part 1/2 of Jorrit Poelen on “How Big is that Bee? On Distributed Publishing of Existing Digitized Specimen Records and Their Derived Knowledge” at Nanosessions #1. See also nanosessions-presentation-2023-02-28.md and hash://sha256/e537ca09b318fe2f314cdda85f7e166d7ea360902cf5724fc41acc0d7e48015d.
28 March 2023 Part 2/2 of Jorrit Poelen on “How Big is that Bee? On Distributed Publishing of Existing Digitized Specimen Records and Their Derived Knowledge” at Nanosessions #2. See also nanosessions-presentation-2023-03-28.md and hash://sha256/ae046add7e565b9e72e69e9fafad8d615ef75d0e77c842e1bf135ba4f5f06704.
30 March 2023 Publication of: Brianne Du Clos, Julien Brun, Sam Droege, Jen Hammock, Andrew Johnston, Jonathan Koch, Clare Maffei, Cynthia Sims Parr, Jorrit H. Poelen, Katja C. Seltmann, & Erika M. Tucker. (2023, March 30). US National Native Bee Monitoring RCN Data Management Workshop: Public Domain Videos (1.0.0). Zenodo. doi:10.5281/zenodo.7868217.
4 December 2023 Symbiota Support Group talk “Image Tagging in Symbiota Portals” by Colleen Smith of the UC Santa Barbara Cheadle Center for Biodiversity and Ecological Restoration. streaming video.
5 January 2024 Seltmann, K., & Poelen, J. (2024). Tab and comma delimited versions of Discover Life bee species guide and world checklist (Hymenoptera: Apoidea: Anthophila) (56.1) [Data set]. Zenodo. https://doi.org/10.5281/zenodo.10019874.
2 February 2024 Seltmann, K. (2024). Exhibit at California Nature Art Museum, San Luis Obispo, CA: the The Birds and the Bees and More: Pollinators hash://md5/4f552ae78473131eafb39657f95c6260. Zenodo. doi:10.5281/zenodo.10612006 .
3 July 2024 Seltmann, K. C., Poelen, J. H., & Global Biotic Interaction Community. (2024). Global Bee Interaction Data (v4.0) [Data set]. Zenodo. https://doi.org/10.5281/zenodo.12639658 .
Collections Status
Click on badges to browse/download indexed records or inspect automated reviews.
ASU /
CAS /
FSCA /
MCZ /
LACM /
SDNHM /
UCB /
UCSB /
UCM /
KU /
UMMZ /
UNH /
| status | institution/collection | platform | contact |
|---|---|---|---|
 |
ASU / Arizona State University ASUHIC / Hasbrouck Insect Collection | Symbiota Light-Ecdysis | Sangmi Lee |
 |
CAS / California Academy of Sciences ENT / Entomology | Symbiota Monarch | Christopher Grinter |
 |
FSCA / Florida State Collection of Arthropods / Entomology | Symbiota Light-Ecdysis | Elijah J. Talamas |
 |
MCZ / Museum of Comparative Zoology, Harvard University ENT / Entomology | MCZbase-Arctos | Naomi E. Pierce, Crystal Maier & Paul Morris |
 |
LACM / Natural History Museum of Los Angeles County LACMEC / Entomology | KEmu | Brian V. Brown & Giar-Ann Kung |
 |
SDNHM / San Diego Natural History Museum SDMC / Entomology Department | Specify | Pamela Horsley & Michael Wall |
 |
UCB / University of California Berkeley EMEC / Essig Museum of Entomology | in-house MySQL | Peter Oboyski & Neil D. Tsutsui |
 |
UCSB / University of California Santa Barbara IZC / Invertebrate Zoology Collection | Symbiota Light-Ecdysis | Katja C. Seltmann |
 |
UCM / University of Colorado Museum of Natural History UCMC / Entomology Collection | Specify | Adrian Carper |
 |
KU / University of Kansas Natural History Museum SEMC / Entomology Division | Specify | Michael S. Engel & Victor H. Gonzalez |
 |
UMMZ / University of Michigan Museum of Zoology UMMZI / Division of Insects | Specify | L. Lacey Knowles |
 |
UNH / University of New Hampshire UNHC / Donald S. Chandler Entomological Collection | TaxonWorks | Istvan Miko |
Integration Profiles
Integration profiles are descriptions on how data flows from one system to the next. Because the Big Bee community is using a variety of tools (e.g., Arctos, Symbiota, Specify, Excel, MSAccess) to manage and share collection data, there are many integration profiles to exchange collection data. The integration profiles below are geared towards the exchange and linking of biotic associations data (e.g., species interactions, or host-parasite associations) with GloBI.
Arctos
Arctos managed collections are indexed by GloBI. This section explains how.
| Arctos | GloBI Integration Profile |
|---|---|
| authors | Dusty (Arctos dev), Mariel Campbell (MSB), Teresa Mayfield-Meyer (MSB) |
| actors | Arctos, VertNet, GloBI |
| integration method | A collection manager uses Arctos to establish associations or relationships between records. Arctos periodically shares data with VertNet. VertNet uses GBIF IPT software to publish data archives. VertNet publishes a list of available datasets in the form of a RSS feed, including those shared by Arctos. Periodically, GloBI finds, and downloads, Arctos related data archives in VertNet. Then, GloBI indexes the associatedOccurrences fields of records in these Arctos data archives. The associatedOccurrences contain the association type (e.g., “eats”) and a pointer to the occurrence id of the linked record. |
| diagram | |
| example collection | MSB-PARA |
Symbiota
Various Symbiota-based collections are indexed by GloBI. This section explains how.
| Symbiota | GloBI Integration Profile |
|---|---|
| authors | Katja Seltmann (UCSB) |
| actors | Collection Manager, Symbiota CMS1, Symbiota Portal1, GloBI |
| integration method | A collection manager uses the “associatedTaxa” fields in Symbiota CMS to record host-parasite associations. The Symbiota CMS periodically publishes their data to a Symbiota Portal (e.g., https://scan-bugs.org , ) . After successful publication, the Symbiota Portal includes the updates data archive in their list of available datasets through their RSS feed. GloBI indexes all data archives in the list of available datasets. For each dataset, GloBI looks for association records in associatedTaxa, associatedOccurrences, dynamicProperties field as well as Resource Relationship and Associated Taxa Extensions. In this case, only associatedTaxa fields are encountered and related records are indexed accordingly. |
| diagram | |
| example collection | UCSB-IZC |
1Note that Symbiota includes two distinct systems: Symbiota CMS and Symbiota Portal. Symbiota CMS is a collection management system typically used to keep a digital inventory of physical collections. Symbiota Portal exposes digital records as Darwin Core Archives and Ecological Metadata Language files, These Darwin Core Archives and EML files are included in a list of available datasets in the form of a RSS feed. So, This include records retrieved from Symbiota CMS as well as other systems or datasources (e.g., spreadsheets, csv files). Symbiota Portal is used to help interface with national and global aggregators like iDigBio and GBIF.
Specify
Association records in Specify managed collections are indexed by GloBI. This section explains how.
| Specify | GloBI Integration Profile |
|---|---|
| authors | Ralph Holzenthal (UMSP), Robin Thomson (UMSP) |
| actors | Collection Manager, Excel, Specify6, https://scan-bugs.org |
| integration method | Collection Manager enters records in excel, then uploads the records in batch into Specify. A manual export to https://scan-bugs.org is done periodically to provide updates to GBIF, iDigBio and GloBI. If updates are made to existing Specify records, a new batch export is needed to update https://scan-bugs.org . |
| diagram | |
| example collection | |
| open questions | 1. Which darwin core archive field to use for assocations? associatedTaxa, associatedOccurrences, Resource Relationship extension. 2. Can we automate the export of Specify records to SCAN? |
| references | Linking Specify data to SCAN Collection Provided by Laura Prado, U. of Wisconsin (December 8, 2018) https://scan-all-bugs.org/?page_id=2084 |
| Specify | GloBI Integration Profile |
|---|---|
| authors | Erika Tucker (UMMZ), Barry Oconner (UMMZ) |
| actors | Collection Manager, Specify, https://gbif.org/ |
| integration method | Collection Manager enters records in Specify. A manual csv export of full Specify database (including host records) is shared with, and indexed by, GloBI. If updates are made to existing Specify records, a manual export performed and GloBI is notified. |
| diagram | |
| example collection | |
| open questions | 1. Which darwin core archive field to use for assocations? associatedTaxa, associatedOccurrences, Resource Relationship extension. 2. How to establish an automated the export of Specify records via (UMich) IPT? 3. How to best link to individual specimen records? 4. How to best cite specimen records? 5. How to establish reliable links to non-UMMZI host records? |
| references | 2020-01-23 Meeting notes and Jan 2020 email exchanges between Jorrit (GloBI), Erika (UMMZ), Barry (UMMZ) |
| example collection | UMMZ, UMMZI on GloBI and UMMZI on github . |
EMu
The EMu <> GloBI integration profiles describes a way GloBI indexes assocation data originating from EMu .
| EMu/GBIF | GloBI Integration Profile |
|---|---|
| authors | Kate Webbink, Janeen Jones |
| actors | FMNH Collection Manager, EMu, IPT, GloBI |
| integration method | A collection manager uses EMu to establish associations or relationships between Catalogue (occurrence) records. The collection manager periodically exports datasets from EMu, and the IT Department publishes those datasets as resources on the FMNH IPT - fmipt.fieldmuseum.org. Datasets (“IPT resources”) that include interactions among occurrences will include a Darwin Core “Resource Relationship” extension. The fmipt site publishes a list of available IPT resources in the form of a RSS feed (https://fmipt.fieldmuseum.org/ipt/rss.do), similarly to VertNet. Periodically, GloBI could find and download FMNH-related data archives in the fmipt. Then, GloBI could index the resourceID fields of records in these FMNH data archives (IPT resources). The relationshipOfResource field contains the relationship type (e.g., “stomach contents of”) and the relatedResourceID field contains a pointer to the occurrenceID of the linked record. |
| diagram | A workflow using a public EMu website for GloBI to link back into: 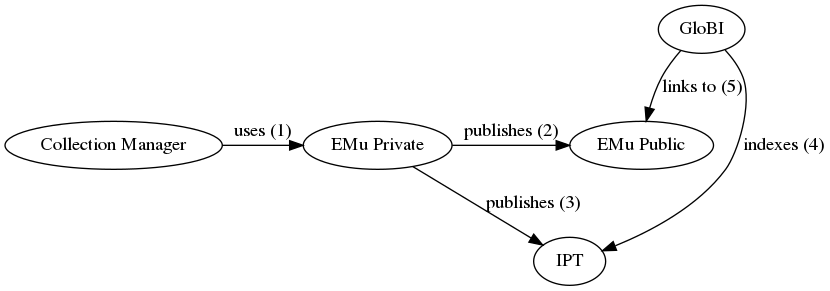 An alternative workflow using a Symbiota Portal for public linking: An alternative workflow using a Symbiota Portal for public linking: 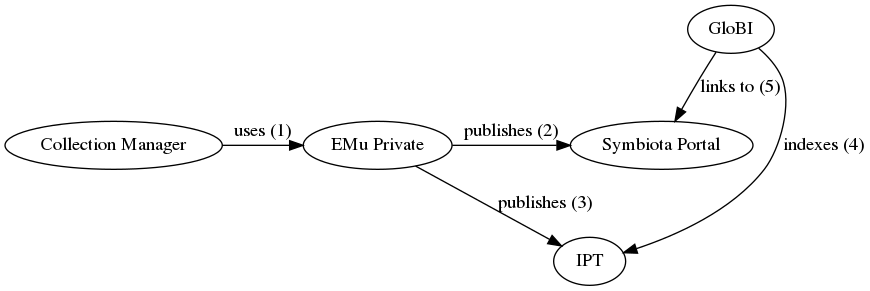 |
| example collection | https://fmipt.fieldmuseum.org/ipt/manage/resource?r=fmnh-rr-test ( https://github.com/jhpoelen/eol-globi-data/files/3586074/dwca-fmnh-rr-test-v1.1.zip ) Post-FMNH/EMu-data-standardization, more resources on fmipt will include a “Resource Relationship” DwC extension |
TaxonWorks
| PSUC | GloBI Integration Profile |
|---|---|
| authors | Andrew R. Deans, Laura Porturas |
| actors | Curator, Data Manager, TaxonWorks, SCAN, GloBI |
| integration method | A curator uses TaxonWorks to manage collection object associations. The data manager periodically exports data from TaxonWorks to SCAN as Darwin Core Archive. Periodically, GloBI indexes PSUC datasets as part of indexing all of SCAN. |
| diagram | ? |
| example collection | ? |
Institutional
Undecided
| Your platform/project here | GloBI Integration Profile |
|---|---|
| authors | ? |
| actors | ? |
| integration method | ? |
| diagram | ? |
| example collection | ? |
Aside from various collection management systems, some collections use custom, home grown tools to manage their digital inventory. When feasible, we aim to develop custom integration profiles for those institutional solutions, provided that the data is openly accessible on the internet.
DwCA Guidelines
Darwin Core Archives provide several ways to capture biotic associations including: associatedTaxa, associatedOccurrences, ResourceRelationship, and dynamicProperties.
For specific examples also see GloBI blog posts Associating with Natural History Collections and Models in Fashion.
General Guidelines
The interaction information should be shared in multiple ways through DwC-A because no single way is sufficient for all aggregators. GBIf does display Dynamic Properties and Associated Taxa (according to K. Seltmann).
Associated Taxa
http://rs.tdwg.org/dwc/terms/associatedTaxa should always be filled out when an association is present (according to K. Seltmann).
Associated Occurrences
If http://rs.tdwg.org/dwc/terms/associatedOccurrences are listed, those should be a separate column in the DwC-A (not included in http://rs.tdwg.org/dwc/terms/dynamicProperties) (according to K. Seltmann).
http://rs.tdwg.org/dwc/terms/associatedOrganisms may include a relationship and link.
Dynamic Properties
http://rs.tdwg.org/dwc/terms/dynamicProperties should be used to handle all other information about an interaction formatted in JSON (according to K. Seltmann).
Resource Relations
https://dwc.tdwg.org/terms/#resourcerelationship is another way of adding biotic interactions to the Darwin Core archive, but only accommodates key:values, thus dynamicProperties will also be needed for more complex information (according to K. Seltmann).
https://dwc.tdwg.org/terms/#resourcerelationship is designed to link, or relate, occurrence/taxon or any kind of DwC record using a defined relationship. This means that many details from the linked records are inherited (e.g., lifestage of a specimen described by a linked occurrence record). However, limited information (e.g., https://dwc.tdwg.org/terms/#dwc:relationshipAccordingTo, https://dwc.tdwg.org/terms/#dwc:relationshipEstablishedDate, https://dwc.tdwg.org/terms/#dwc:relationshipRemarks) can be associated to the relationship record itself (according to J.H. Poelen).
(work in progress) Include best practices, pros/cons, examples and links to datasets using the extensions.
Interaction Types
Supported Terms
Translation Tables
GloBI uses a subset of biotic interaction (or association) terms defined in the OBO Relations Ontology (OBO RO) to help classify and index biotic associations in collection records. Verbatim association types (e.g., on, parasite of, found on) are explicitly mapped into these OBO RO terms using translation tables. GloBI keeps a default translation tables and specific collection may choose to provide their own (see e.g., INHS-Insects ).
| resource | description |
|---|---|
| OBO Relations Ontology project page | OBO RO contains many kinds of terms, not just biotic associations terms |
| List of OBO RO Biotic Interaction Terms with definitions | a table of RO biotic interaction terms and their definitions (if available) |
| List of GloBI Supported Interaction Terms | subset of RO interactions terms that GloBI uses for indexing |
| Default Verbatim Terms Translation Table | the translation table used by GloBI to maps verbatim interaction terms to supported interaction terms |
| Example of Custom Verbatim Terms Translation Table | if provided/needed, GloBI can use a custom mapping provided by a collection |
The OBO RO is far from complete and we expect to add new terms and improve definitions as needed. Also, GloBI translation tables can be easily updated when needed. Please open an issue if you have questions or suggestions.



